Avoid mitochondrial toxicity affecting the integrity and dynamics of mitochondria
Mitochondria are dynamic organelles with a complex structure of membranes and inner compartments. The cell’s central metabolic hub is organised as a mitochondrial network, the morphology of which results from a delicate balance between fusion and fission, biogenesis, and elimination of altered mitochondria by mitophagy. Our technological platform MiToxView® provides custom-designed assays to investigate mitochondrial integrity and dynamics in whole cells and mitochondria from various sources.
The stress-response to mitochondrial toxicants can involve the modification of mitochondrial dynamics leading to disorganisation of the network and mitochondrial dysfunction.
In addition, some compounds can affect mitochondrial integrity, either specifically by inducing mitochondrial membrane permeabilisation (MMP) by opening mPTP, or through a detergent-like non-specific effect. The ensuing loss of transmembrane potential and decreased ATP production trigger cell death and organ toxicity.
Some drugs, such as fialuridine, can also alter and/or deplete mitochondrial DNA, which encodes for 13 proteins of the respiratory chain. This type of damage induces dysfunction in electron transfer through the respiratory chain potentially leading to metabolic dysfunction.
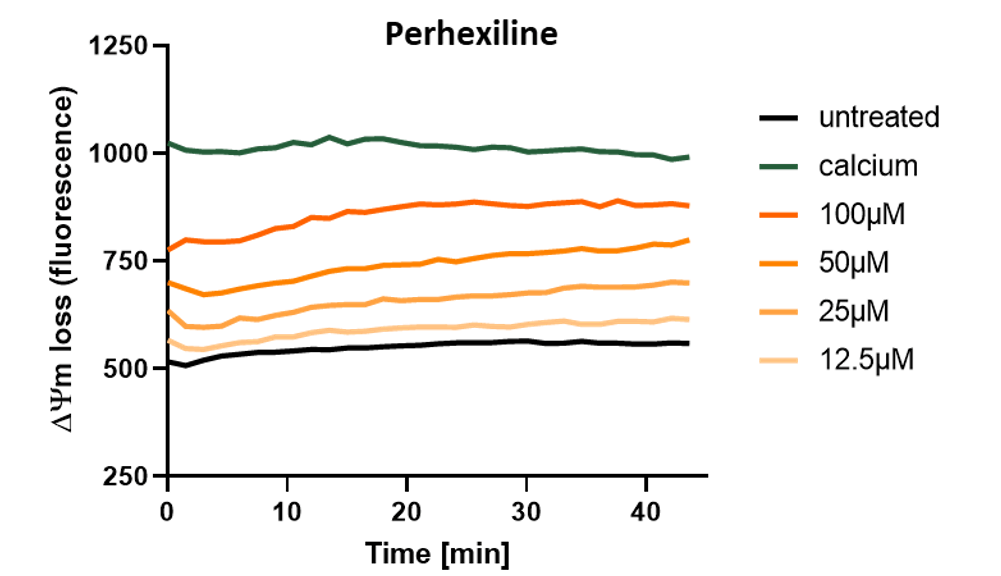
Measure transmembrane potential loss on isolated mitochondria as an indicator of direct mitochondrial toxicity of compound.
Induction of transmembrane potential (ΔΨm) loss on isolated mouse liver mitochondria by acute treatment with Perhexiline showing direct mitochondrial toxicity of this hepatotoxic drug. Compound dose-response is monitored by spectrofluorimetry during 45 min in presence of Rhodamine123.
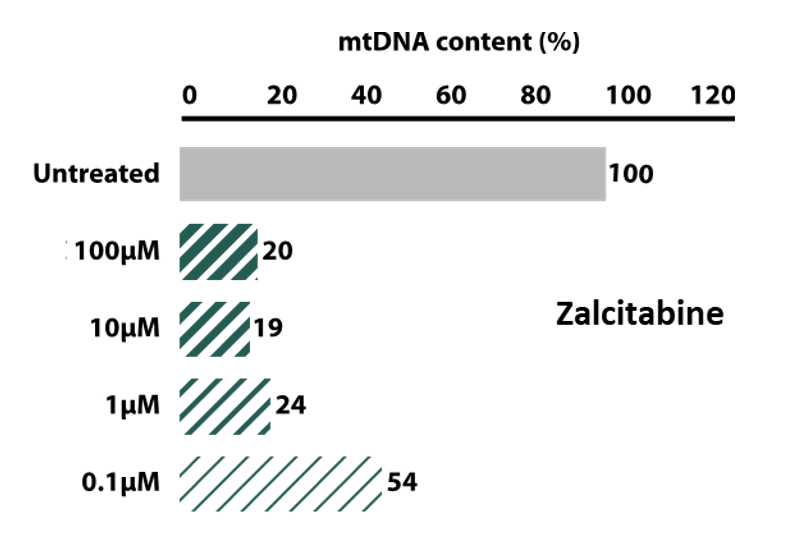
Quantify mtDNA depletion that leads to mitochondrial respiratory chain dysfunction (mechanism of action of some nucleoside analogs in hepatotoxicity).
Mitochondrial toxicity of the anti-retroviral compound Zalcitabine (ddc) elicited by quantification of mtDNA after long-term and chronic treatment of differentiated HepaRG® cells.
Our platform provides consolidated data to help you to:
- Determine potential mitochondrial toxicity of compounds
- Screen and rank compounds based on their effects on mitochondrial integrity: mPTP opening, mtDNA depletion, etc.
- Understand the mitochondrial toxicity mechanism induced by a compound: effect on mitochondrial membrane integrity, transmembrane potential, dynamics, etc.
- Evaluate the impact of a compound’s mitochondrial toxicity at the cellular level: fusion, fission, mitophagy, etc.
- Predict mitochondrial toxicity in humans: hepatotoxicity/DILI, cardiotoxicity, etc.
- Analyse mitochondrial damage in pre-clinical and clinical biological samples.
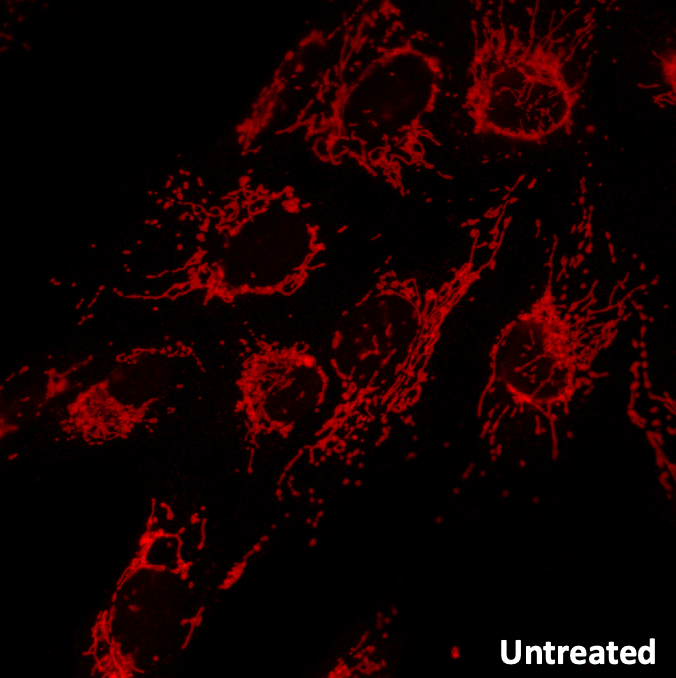
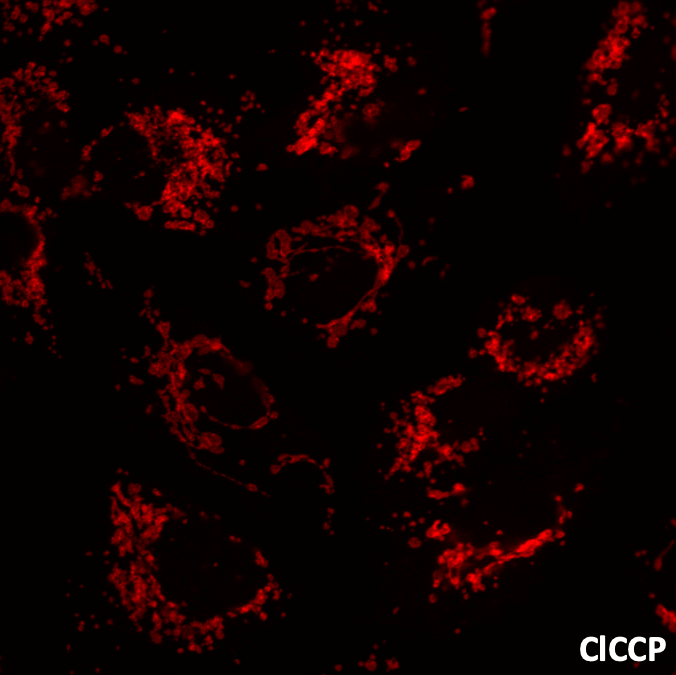
Investigate the fission–fusion balance of mitochondrial network as a marker of mitochondrial toxicity.
Alteration of mitochondrial network (fission) in non-neoplastic human hepatocyte HHL-16 cells before and after treatment with the uncoupling agent carbonylcyanide-3-chlorophenylhydrazone (CCCP). Fluorescent microscopy is performed after labeling with anti-Tom20 antibody.